Accelerating genomics variant interpretation with AWS HealthOmics and Amazon Bedrock AgentCore
aws.amazon.com - machine-learningGenomic research stands at a transformative crossroads where the exponential growth of sequencing data demands equally sophisticated analytical capabilities. According to the 1000 Genomes Project, a typical human genome differs from the reference at 4.1–5.0 million sites, with most variants being SNPs and short indels. These variants, when aggregated across individuals, contribute to differences in disease susceptibility captured through polygenic risk scores (PRS). Genomic analysis workflows struggle to translate such large-scale variant data into actionable insights. They remain fragmented, requiring researchers to manually orchestrate complex pipelines involving variant annotation, quality filtering, and integration with external databases such as ClinVar.
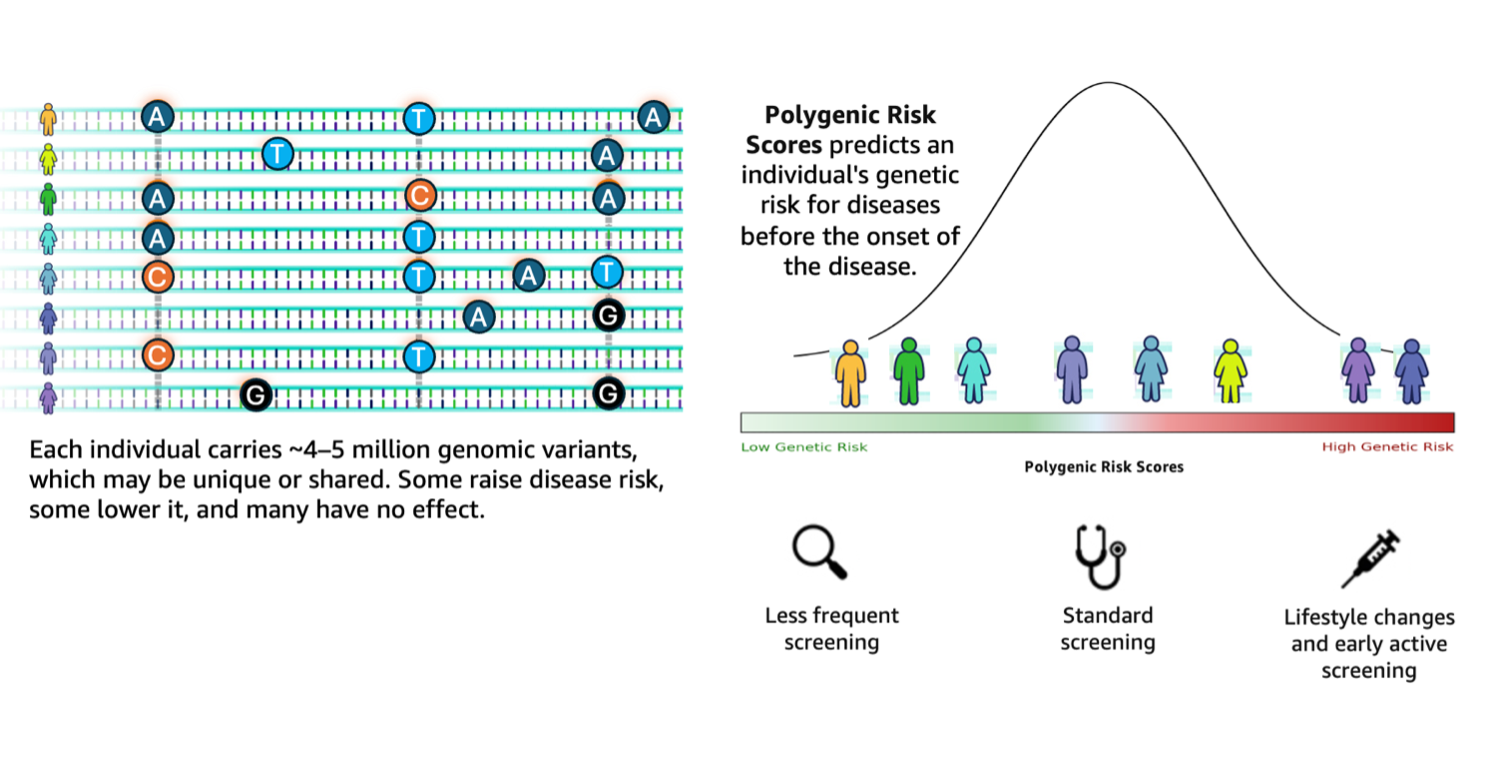
AWS HealthOmics workflows along with Amazon S3 tables and Amazon Bedrock AgentCore together provide a transformative solution to these challenges. HealthOmics workflows support the seamless integration of annotating Variant Call Format (VCF) files with insightful ontologies. Subsequently, the VEP-annotated VCF files need to be transformed into structured datasets ...
Copyright of this story solely belongs to aws.amazon.com - machine-learning . To see the full text click HERE



